What is ChIP-seq? ATAC-seq?
What is ChIP-seq?
ChIP-seq is an experiment that answers the question: is a protein bound to a piece of DNA or not? Scientists use “ChIP-seq” to find out if a “protein of interest” is bound to a piece of DNA or not. Usually the protein of interest is a “transcription factor”.
What is a transcription factor?
A transcription factor is a protein that binds to DNA, and starts the process of “transcription” - the conversion of DNA to RNA. The human genome has about 2,600 transcription factors (TF’s). What makes a protein a TF is if it has a ‘DNA binding domain’.
What is the point of transcription?
Transcription is the first step of “gene expression” where a gene is converted into a functional product usually this “product” is a protein, but sometimes the “product” is functional RNA – such as transfer RNA (tRNA) or small nuclear RNA (snRNA). In humans, genes are 1,148 to 37.7 kilobases (kb) long, with the average length of a gene being 8,446 base pairs.
What is the point of making mRNA?
Three letters of mRNA is called a ‘codon’, and codons code for amino acids. There are 20 naturally occurring amino acids, and chains of amino acids together form “polypeptides” or informally, proteins. The process of converting mRNA into proteins is called “translation”.
A Codon to Amino Acid Conversion Table
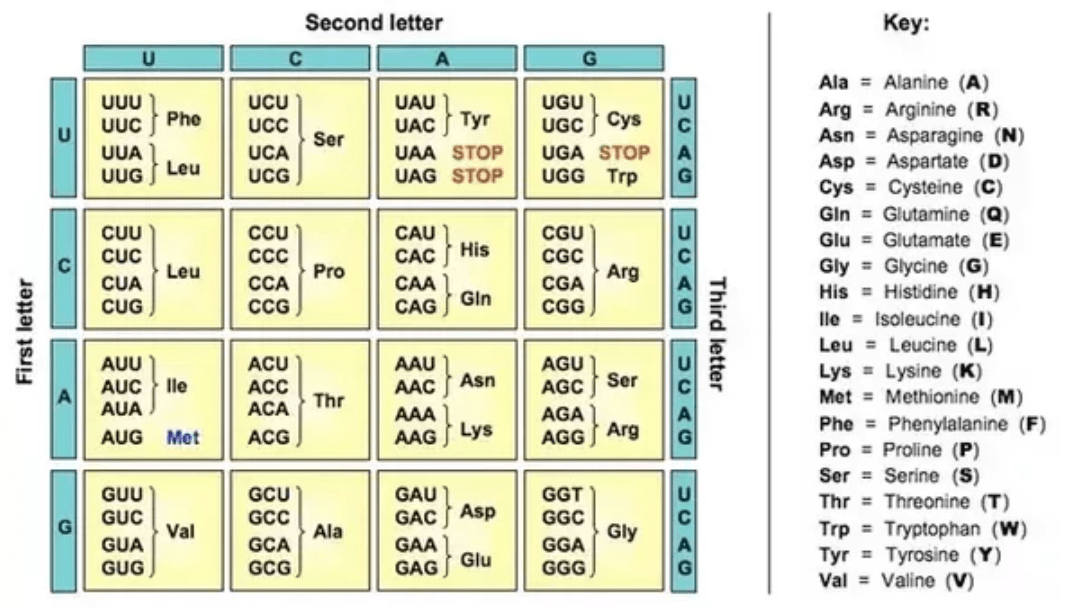
There are 4 mRNA bases (U, C, A, and G), and a codon is 3 bases, so we see that there are 4 options for each position, so 4x4x4 = 64 different combinations of codons. These 64 codons code for 20 amino acids, so on average every amino acid has 3 codons that code for it. Some amino acids, like Arginine, have 6 codons that code for it. This is called the “degeneracy of codons” (the fact that each amino acid has multiple codons that code for it) - which is a feature rather than a bug because the redundancy means that if a DNA base is mutated (damaged, or copied incorrectly) the amino acid produced can still be correct, meaning that the downstream protein can still be functional.
This is a good review of how transcription and translation works.
TLDR;
- Transcription - DNA gets converted into RNA
- Translation - mRNA gets converted into proteins
Back to ChIP-seq!
“ChIP-seq” is short for “chromatin immunoprecipitation (ChIP) combined with DNA sequencing”.
How does ChIP-seq work exactly? What is the experimental procedure?
Goal: Discover what sequence(s) of DNA a protein binds to
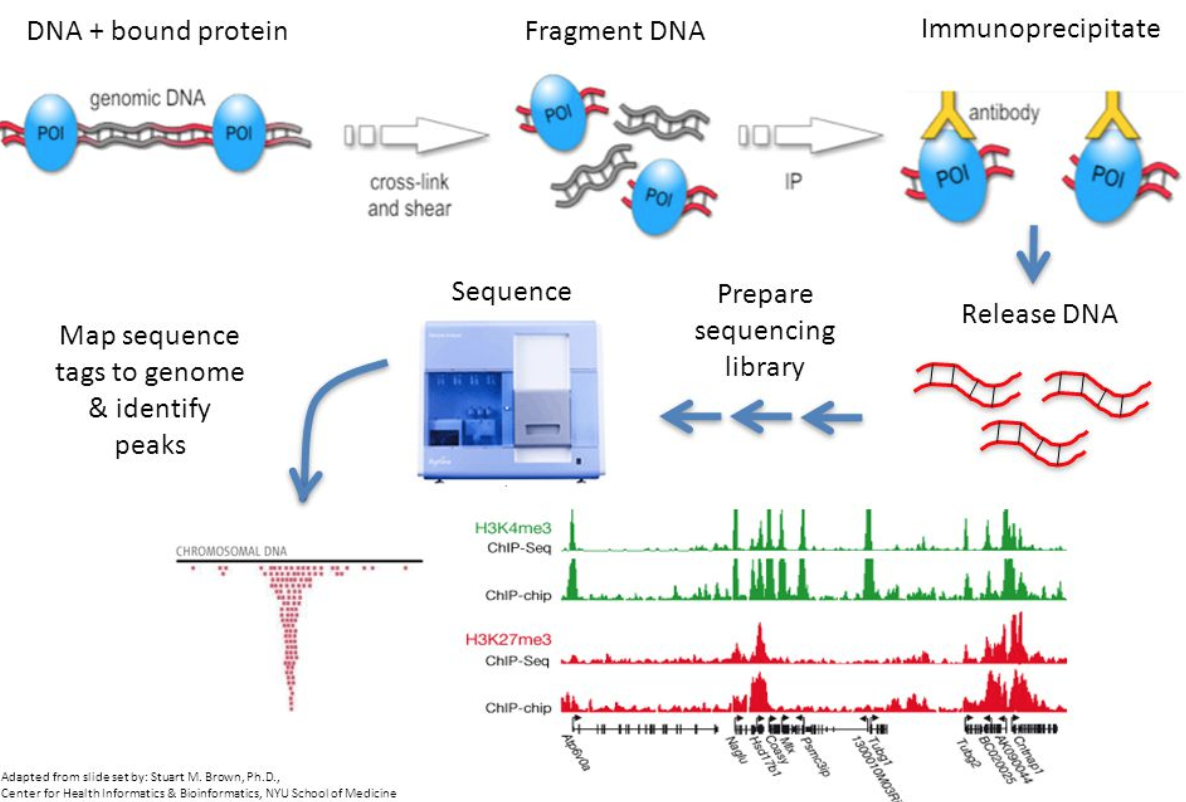
- break up the DNA into small pieces, around 100 base pairs in length (sonication)
- wash the DNA with the target protein. The protein will bind to specific sequences of DNA. (enrichment)
- use antibodies that bind specifically to the target protein to grab the DNA that has the protein attached to it
- make more copies of the DNA (amplification) with the protein attached to it
- sequence the DNA
What is ATAC-seq?
ATAC-seq is an experiment that answers the question: is the DNA in a region of open chromatin or closed chromatin?
-
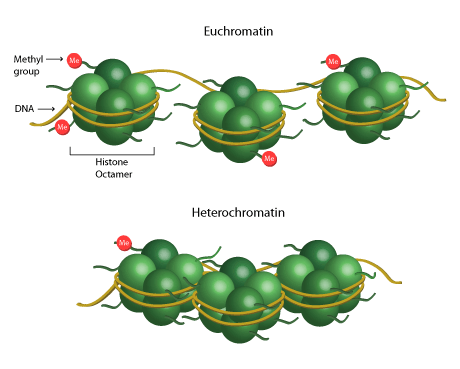
Open chromatin is called “euchromatin”, which is pictured on top. The DNA is the yellow rope. Notice that the DNA is accessible here! Transcription factors and other proteins are able to bind to the DNA since it is exposed (not wound around the histone).
-
Closed chromatin is called “heterochromatin”, which is pictured on the bottom. The DNA is not accessible since it is tightly wrapped around the histone octamer.
ATAC-seq uses an engineered Tn5 transposase to cleave DNA and to integrate primer DNA sequences into the cleaved genomic DNA (that is, tagmentation)
DNase-seq and FAIRE–seq are also used to determine if a piece of DNA is in open or closed chromatin.
- DNase-seq - chromatin is lightly digested by the DNase I endonuclease. Size selection is used to enrich for fragments that are produced in regions of chromatin where the DNA is highly sensitive to DNase I attack
- FAIRE-seq - formaldehyde is used to crosslink chromatin, and phenol–chloroform is used to isolate sheared DNA.
Still curious?
Videos
- StatQuest: A gentle introduction to RNA-seq
- StatQuest: A gentle introduction to ChIP-Seq
- An introduction to ChIP-seq analysis
Academic Journals